Part 1 of this article started with the Hominina / Paninae (chimpanzee) split of about 6.5 mya and ended with the emergence of H. heidelbergensis about 850,000 ya. Part 2 will look at some of the Homo species that have roamed the earth from then until the present, which sees just H. sapiens representing the Hominin line. Figure 1 provides a high-level overview of the species covered in both parts.
DNA sequencing technology, specifically ancient DNA, has advanced tremendously recently, and this has greatly elevated the scope and sophistication of the study of Hominin species. I had planned on making DNA sequencing a major part of this article, but quickly realised that it is too large and complex a topic to adequately cover, and so that will have to wait for a future Science Break article. In the abbreviated section below on DNA sequencing I draw heavily on Svante Pääbo’s Neanderthal man: in search of lost genomes, a book that I highly recommend to readers who want to pursue this topic further. Pääbo has played a central role in the push to salvage, reconstruct and sequence archaic DNA, and very effectively explains the many challenges and clever solutions that have allowed us to probe the genetic relationships between ourselves and ancient ancestors. In addition to being a brilliant scientist, Pääbo is a very interesting character, so his book is well worth the read.
DNA Sequencing
The concept of using genetic markers to study the history and relationships of human population groups is not new. Beginning in the 1950’s blood types were used to identify distinct human populations with a common heritage. That led to the use of protein markers, and then in the 1980’s the development of polymer chain reaction (PCR) technology resulted in the concept of evolutionary distance between alleles. Various methodologies thus matured during this period, and were ready and waiting when DNA sequencing exploded onto the scene. As with any field of science, it has its own unique methods and language, and papers can be quite difficult to interpret and understand. There is extensive use of statistics, and this can be daunting to the non-expert.
The factors working against the preservation of DNA are numerous and formidable. Even while we are alive, our DNA is constantly being attacked and degraded, and our bodies contain several repair systems to maintain the integrity of our DNA. The double helical DNA molecule is built from four nucleotides: adenine, thymine, guanine and cytosine; A, T, G and C in short. Adenine (A) nucleotides in one strand form pairs with T’s in the other, and G and C pair off in a similar fashion. The patterns and sequences of these pairs make up the encoded blueprints for our bodies. C naturally degrades into uracil (U), so enzymes must continually gather U’s and replace them with C’s. Other enzymes replace random lost nucleotides that create breaks in our DNA. These and many other molecular goings on are happening all the time, and if not repaired it is estimated that our genomes would be rendered useless within hours.
With death most, if not all, of the body’s repair systems stop working properly. The membrane walls that compartmentalize the various repair and defense enzymes begin to break down, allowing enzymes to carry out actions willy-nilly, such as cutting and splicing (see (Kuhn, CRISPR Genome Editing, 2016)). With time bacterial degradation also comes into play, as well as destructive radiation from space. Counteracting all these factors working against the preservation of DNA is the fact that our bodies are made of about 1 trillion cells, and each cell contains our complete DNA. Most of the degrading processes require water. The high number of DNA molecules present in each organism, if fortuitously combined with a dry environment, results in a good chance of some preservation, but the DNA preserved is likely to be a jumble of unidentifiable chopped up bits. This is the challenge that faced scientists like Pääbo who began to tackle it in the early 1990’s, akin to solving a very complicated jigsaw puzzle, with 6.4 billion nucleotide pairs making up the full human genome.
After spending thousands of years encased in cave dirt, ancient Hominin bones are usually extensively touched by many humans as they are cleaned in the field, transported to some university to be handled, viewed, compared, identified, and catologued, as well as often being subjected to cleaners and glues that can contain organic components. By the time small amounts of bone marrow are extracted, the samples are usually highly contaminated by foreign DNA, human and other. What follows is painstaking work involving numerous steps to isolate, filter, capture and bind the ancient DNA fragments for analysis.
PCR techniques are critical to DNA sequencing. For example, PCR is used to duplicate a single piece of archaic DNA billions of times over, providing a rich, redundant data set to work on. The input DNA is heated in a mixture containing the source DNA, an enzyme called polymerase DNA, DNA primers, and the four nucleotide building blocks, A, T, G and C. As the mixture warms, the two helical strands of the input DNA segment split apart. The synthetic DNA primers then bind to specific start and end points on the targeted DNA. As the mixture cools, the DNA polymerase enzymes go to work, duplicating two new strands using the raw nucleotides. Each additional cycle sees a doubling of new strands, so exponential cloning is achieved.
The actual identification and registry of the various nucleotides is now automated. Many methods exist, but I believe most are based on pyrosequencing, a method that builds on PCR cloning. In an early version, the Sanger method, a small portion of the nucleotides were modified in two ways: each of the four types was given a unique fluorescent colour, and they were designed so that when they were used by the enzyme to build a pair, the synthesis stopped. The mixture then contained, along with other stuff, synthesized pieces of four distinct and different lengths, and the last pair of each of these lengths was uniquely identified by its colour. The process was then repeated, and labouriously the sequence was built up, four pairs at a time.
Obviously, the Sanger method was very slow, and a breakthrough came when Swedish biochemist Mathias Uhlén cleverly exploited an idea developed by fellow Swede Pål Nyrén. He adds only a single nucleotide to the mix each cycle, again incorporating a fluorescent dye to that single type of nucleotide. Each time the enzyme incorporates the dyed nucleotide, it gives off a flash of light, which is detected by a camera, and its location is catalogued. The cycle rips through the entire DNA sequence, and then the next cycle does the same, but with a different nucleotide. Thus in four cycles, the entire sequence is very quickly and automatically sequenced. There are now many different modern methods of sequencing, each with its own advantages and disadvantages. Advances are allowing longer and longer pieces of DNA to be sequenced, and faster.
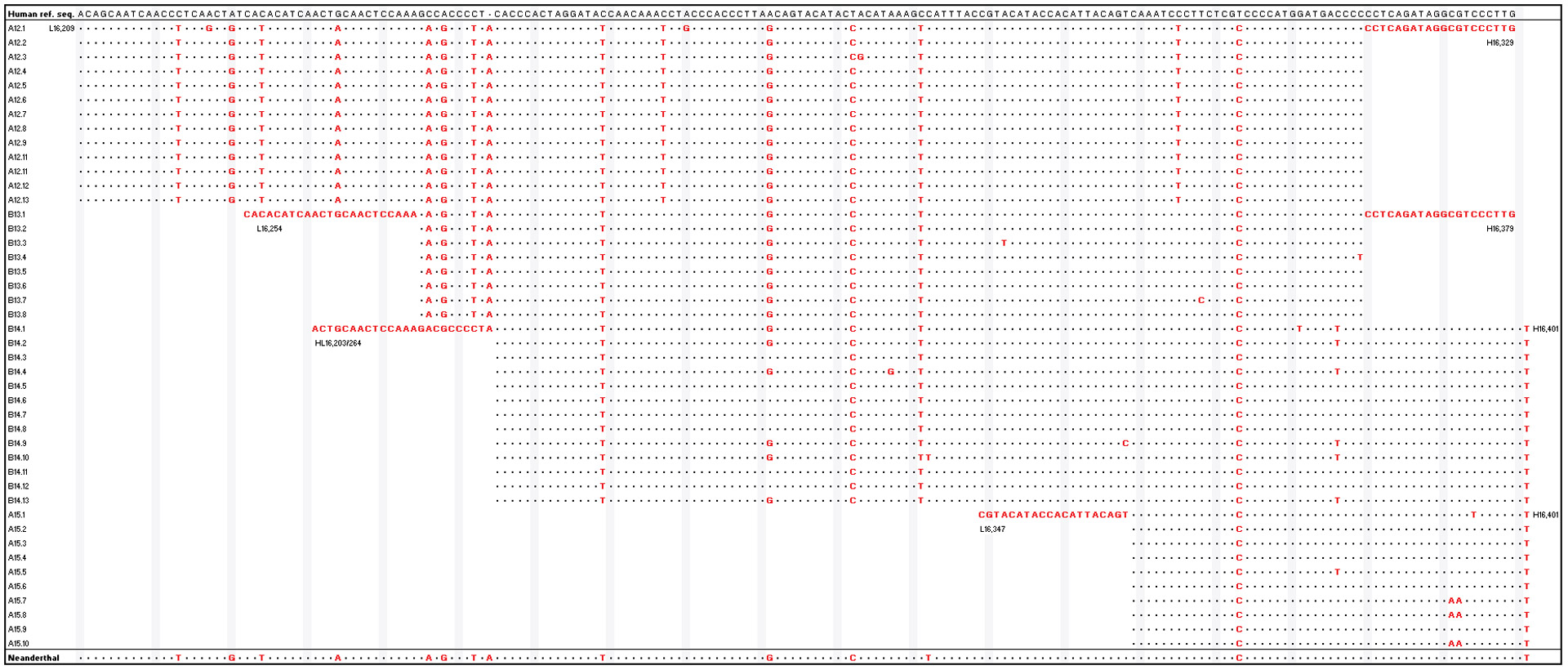
Figure 2 used as an example in Pääbo’s book, shows a section of Neanderthal mtDNA (bottom line) that has been reconstructed by comparing a reference human sequence (top line) against 44 sequences derived from cloned molecules taken from a Neanderthal bone (incidentally, found in the Neander Valley which the species is named after). The dots in the Neanderthal sequence indicate that those nucleotide pairs are the same as the human sequence, and the letters indicate the nucleotides that differ from the human sequence. Ancient Hominin DNA is close enough to human DNA that the little bits can be matched up against modern human DNA to determine their locations – for example, there is only ~0.3% difference between our DNA and that of Neanderthals. Overlaps between the bits provide confidence that placement is correct. Interestingly, the term “fold” is used exactly as we do with seismic data – the amount of overlap available is called fold, and obviously the higher it is the greater the chance of getting placement right, as damaged and degraded nucleotides can be misidentified and are just like the seismic noise we combat with higher fold.
The early DNA work began on the parts of the genome that are uniparental, meaning the mitochondrial mtDNA that is passed intact exclusively mother-to-daughter, and the non-recombining parts of the Y chromosome (NRY) that are similarly passed father-to-son. These uniparental genes have known mutation rates which provide a sort of evolutionary clock, but the genetic drift caused by this mutation coupled with the uniparental aspect make them poor sources of evolutionary information.
Many early conclusions regarding Hominin history were based on the analysis of mtDNA. This is because, first, it is far more common than DNA, with many hundreds of copies of it in each cell (as opposed to the single strands of paternal and maternal DNA), and therefore more likely to be preserved than DNA, and second, it is very short (16,569 base pairs versus ~3.2 billion base pairs in single strand DNA) and therefore much more likley to be found fully intact than ancient DNA. Subsequent work has shown some of the earlier conclusions to have been incorrect. For example, mtDNA indicated that there was no mixing between humans and Neanderthals, and many of us who love a romance were greatly let down by this pronouncement. But then – be still, my beating heart – study of the entire recombining genome (the parts that are passed on 50% from mother, 50% from father) showed this to be incorrect – humans and Neanderthals did interbreed, a lot. There are a number of explanations for this disagreement. For instance, perhaps human-Neanderthal hybrid females tended to be sterile, similar to horse-donkey hybrids. Or perhaps it is a statistical reality that all the family lines involving hybridization that have so far been sequenced at some point experienced at least one generation with no female offspring, thus breaking the mitochondrial chain. I find it hard to wrap my head around these sorts of thought experiments, and over time have stopped paying too much attention to uniparental gene analysis studies, as I feel them to be unreliable, or perhaps a better way to put it would be to say they give a limited perspective and should be taken with a grain of salt.
Currently the oldest genome to have been sequenced is mtDNA from 400,000-year-old H. heidelbergensis bones found at Sima de los Huesos, northern Spain. Sequencing the genomes of Hominin species, and then establishing percentages of one species’ DNA found in another’s, tells us a lot. It obviously means the two species had to have overlapped in time and space, gives estimates of when that interbreeding occurred, and how much interbreeding occurred. On average, all non-sub-Saharan humans possess 1.5-2.1% Neanderthal DNA; this number is higher in people of European and Chinese origin, about 2.8%. The DNA of people from Oceania (Papua New Guinea, Bougainville Island, and Australian Aboriginals) is 4-6% Denisovan. Reading this and knowing where Neanderthal and Denisovan bones have been found, one immediately wants to make sense of this, and speculations about the possible movements and interactions of these Hominins over time are very intriguing. Far more sophisticated studies are being conducted. For example, DNA analysis is revealing more detail around when humans migrated from Asia to the Americas, where they came from in Asia, and from which Asian populations.
DNA analysis reveals much more about ancient Hominins than their mass movements and interactions. For example, there are indications that non-African humans derived genetic characteristics from the Neanderthals that were advantageous as they left Africa and settled in more northerly latitudes. Neanderthal alleles related to keratin are found in humans, which may have given early Europeans their lighter skin and thicker body hair. Every few months exciting new results involving the analysis of archaic Hominin DNA are released, and I expect these efforts to increase as DNA sequencing methods become cheaper, faster, more accessible, and more effective.
H. Heidelbergensis
There is general agreement that H. heidelbergensis is the common ancestor shared by us, the Neanderthals, and most likely the Denisovans as well. Probably the only major debate is whether H. heidelbergensis really deserves to be a separate species, and there are strong arguments that it should be classified as a variant of H. erectus. This type of argument will never go away, as there is a fundamental difficulty assigning specific names to living entities that gradually change over hundreds of thousands, if not millions of years. H. erectus didn’t enter its cave one night to emerge as H. heidelbergensis the next morning, but taxonomic names are black and white, and therefore paleoanthropologists must debate and pick the best clear dividing lines, because we do need names and some kind of nomenclature to make sense of it all. The gradual changes that are evolution do not lend themselves to naming conventions, which inherently imply sharp boundaries, so we just must live with this situation.
Keeping things at a realistically blurry level, we’ll say that H. heidelbergensis started to split from H. erectus around 800,000 ya, probably in South Africa, and by 700,000 was distinctly different enough to be considered a new species. Around 500,000 ya H. heidelbergensis found its way into Europe and in response to the different challenges faced in this northern climate, evolved into H. neanderthalensis. Around the same time H. heidelbergensis made its way into Central Asia, and this population evolved into H. denisoviensis. The H. heidelbergensis population that remained in Africa continued on its own evolutionary path, resulting in a new species given the name H. rhodesiensis; it is from this species that H. sapiens eventually sprung from around 300,000 ya, but these direct ancestors of ours did not expand beyond Africa until much later, about 100,000 ya or later.
Homo Neanderthalensis
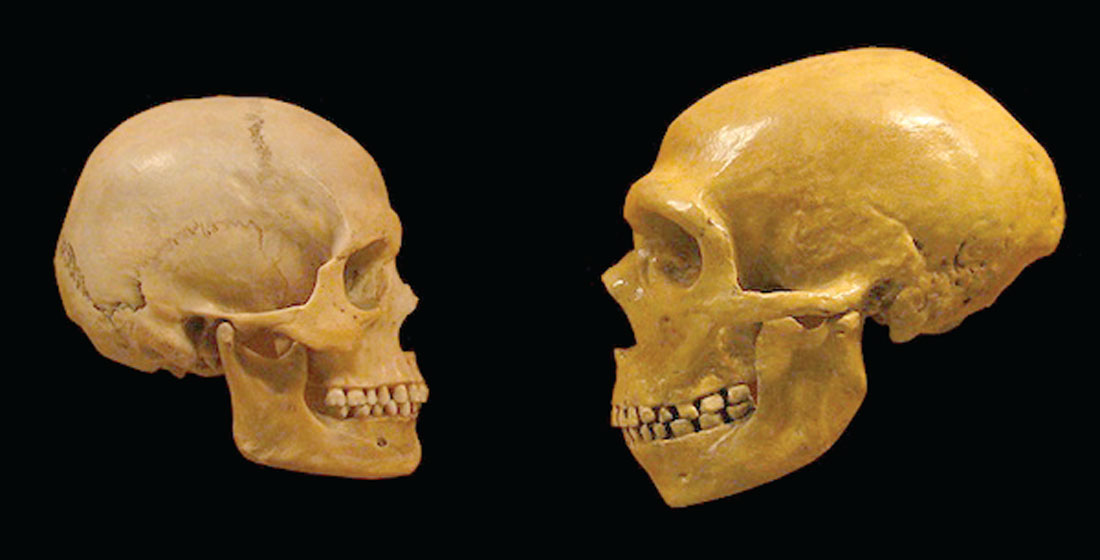
There is a fascination with the Neanderthals among people of European descent, including myself. It probably started when early finds coincided with the 1856 publication of Darwin’s On the Origin of the Species – here was proof of this new theory of evolution, involving our own species, in our own back yard no less! I wrote a short article on Neanderthals in the January 2013 issue of the RECORDER (Kuhn, 2013). Since then many more DNA-based details regarding Neanderthal timing and interbreeding with humans have emerged. Figure 3 shows human and Neanderthal skulls side-by-side. Neanderthals actually had larger brains than us, but recent studies suggest that compared to us, proportionally more of their brains were devoted to vision and physical body control, and less to higher cognitive functions. This would suggest that they were a very physical species, more agile and strong than us, but perhaps not as capable of planning and cooperative behaviors as humans.
The evolution, in Europe and the Middle East, of H. neanderthalensis from H. heidelbergensis began around 500,000 ya and was complete by about 250,000 ya. Neanderthal fossils dated from that point on until about 40,000 ya are found all over Europe, the Middle East, and North, West and Central Asia. During that period, it is believed that there were three main episodes of interbreeding between Neanderthals and humans. The DNA from all non-sub-Saharan humans contain the oldest Neanderthal content, indicating that the first interbreeding happened just when H. sapiens first exited Africa, say around 100,000 ya, probably in the Middle East. Neanderthal DNA content from the second period of interbreeding is not found in Melanesians (for example Australian and Papua New Guinea aboriginals) so they must have split off and moved eastwards prior to this interbreeding. To further spice things up, the Melanesians had relations with Denisovans after this split. The evidence of the last and most recent episode of human-Neanderthal interbreeding is found only in East Asians, so it most likely occurred in East/Central Asia, removed from the Europeans to the west and the Melanesians to the east, and involved human ancestors of today’s East Asians.
Last Christmas I gave my son a DNA test kit as a gift. It showed, predictably, that he has the Northern European genes from my side of the family, and from my wife, predominantly S.E. Asian genes, but also smaller percentages of Melanesian and East Asian DNA markers. This means that he and his sister most likely have bits of DNA from all three episodes of interbreeding with the Neanderthals, plus that of the Denisovan episode.
Homo Denisoviensis
Our knowledge of the Denisovans is based entirely on just five specimens from four individuals – a toe bone, a finger bone, and three teeth. In fact, most of the understanding of their DNA comes from material extracted from the tiny toe bone of a little girl, about half the size of my pinky finger nail – it’s really incredible when you think about it. All five artefacts were found in the Denisova Cave, deep in the Altai Mountains of Russia, about 1000 km directly east of Astana, the capital city of Kazakhstan. Fortunately, the dry cool conditions in this cave are very kind to archaic DNA, and Pääbo and others have been able to achieve full DNA sequencing. The resulting studies have revealed a very complex picture of Hominin migration and interbreeding. A likely scenario is that the pre-Neanderthal/Denisovan population began to split off from H. heidelbergensis in the Middle East approximately 600,000 ya. Around 400,000 ya one group from this population moved off to the NW and into Europe, to become the Neanderthals; a second group moved off to the NE into Asia to become the Denisovans. In the mid-Pleistocene, around 800,000 ya, the glaciation cycles locked onto a period of 100,000 years, conveniently resulting in warm inter-glacial periods that occurred right around the 100,000 year marks, meaning 400,000 ya Asia and Europe were likely warm and attractive places to move into.
While the Neanderthals established their European and near East territory, the Denisovans likely were doing the same in Asia, and probably spread as far as Papua New Guinea. A lot of work has been done comparing the Denisovan genome with those of many different Asian populations, such as the Han, different Melanesian types, Australian Aboriginals, and other aboriginal peoples in Asia, such as the Mamanwa of the Phillippines and the Jehai of Malaysia. All this suggests that the Denisovans were established all across East Asia, and as they moved towards extinction their last domain was in Wallacea (the islands east of the Wallace line, a line running running north from between Bali and Lombok, defined by water deep enough to have represented a barrier even during ice age sea level lows).
At some point Neanderthals pushing east into Central Asia interbred with the Denisovans in the Altai region, and the first wave of modern humans moving eastward – the ancestors of the Melanesians and Australian Aboriginals – also interbred with the Denisovans. It has also been suggested that the Red Deer Cave people (see section below) may have been a recent (~12,000 ya) human-Denisovan hybrid, but no DNA sequencing of their genome has been successful so this remains conjectural. Intriguingly, the Denisovan genome contains components from an unknown Hominin that greatly predates the H. sapiens / H. neanderthalensis / H. denisoviensis split, dating back to a common ancestor from between 1.1 and 4 mya; I wonder if perhaps the Denisovans encountered and interbred with a remnant population of H. erectus in Asia? A lot of really cool genetic studies are being conducted, and coming up with DNA-based theories. For example, the characteristic brown skin and curly hair of Melanesians may have come from the Denisovans, as perhaps did the high altitude oxygen uptake adaptions found in modern day Tibetans.
Homo Rhodesiensis
Meanwhile, back in Africa... H. heidelbergensis was evolving into H. rhodesiensis, although the distinction appears mainly to be based on stone tool improvements around 600,000 ya, and some morphological differences which may or may not distinguish the two. Perhaps the same factors which led to H. heidelbergensis leaving Africa and evolving into the Neanderthals and Denisovans also drove some big steps forward within the population that remained in Africa. Let’s just say H. rhodesiensis went about its business in Africa, and represents an intermediate step between H. heidelbergensis and modern humans, H. sapiens.
Homo Naledi / Homo Cepranensis / Red Deer Cave People
There are a few random fossil Hominin finds that may or may not represent different Hominin species, and I’ll mention three such here. There was a single H. cepranensis skull cap found in 1994, less than an hour’s drive SE of Rome, Italy. Its morphological features are so unique that it is hard not to classify it as a new species. It has been dated at around 450,000 years old, based mainly on stratigraphy. It could therefore have evolved (albeit rather quickly) from early H. heidelbergensis in Europe, or perhaps represents another African Hominin that entered Europe at around the same time. Or perhaps it evolved directly from H. erectus and remained in an isolated Italian pocket for a long time? With a single find, who knows – the possibilities are basically boundless.
H. naledi bones, many of them, were first discovered in just 2015, in the Rising Star cave system 50 km NW of Johannesburg. H. naledi was clearly part of the Homo genus, but definitely not part of the line leading to H. sapiens. Many of its features would suggest that it may have been a descendant of an australopithecine line. Indeed, it was originally assigned a date in the millions of years ago, but the bones have now been accurately dated at between 236,000 and 335,000 ya. This raises the interesting likelihood that it co-habited or alternated occupation of the caves with H. rhodesiensis. So not only were Hominin cousin species like the humans, Denisovans and Neanderthals overlapping, here is an instance of two widely diverged Hominin species living side by side.
Regarding the Red Deer Cave people, no, the folks up the highway [reference to city of Red Deer, AB] have not been identified as a separate Homo species! The name is given to a possibly distinct Hominin species found in caves in Yunnan and Guangxi, China. The bones have been dated to between 14,500 and 11,500 ya, making them the most recently surviving non-human Hominin. They possessed many features that are characteristic of more primitive Hominins such as H. erectus or H. habilis, such as thick skull bones, huge molars, and key to supporters of a H. erectus link, proportionally long femurs. Absent of DNA evidence, theories are floated such as them being a Denisovan-human hybrid, or perhaps the last remnants of H. erectus, hence my earlier speculation that the unknown DNA found in the H. denisoviensis DNA could be from H. erectus. I personally am drawn to theories such as this – I seem to like the idea of species thought to have become extinct long ago actually hanging around much longer in isolated pockets. I suppose it’s similar to the appeal of stories such as King Kong, or perhaps even myths such as the Loch Ness Monster, and I really should be more disciplined and sober when considering these things.
Homo Floresiensis
I know I am not alone in these flights of fancy because the discovery of H. floriensis remains on the Indonesian island of Flores in 2003 certainly captured the imagination of the general public. The diminutive stature of these creatures (the estimated height of adults is 3.5 feet), and the fact that they lived until relatively recently (~60,000 ya) meaning possible overlap with early humans in the region really struck a chord – perhaps legends such as dwarfs and leprechauns have some basis in fact, and distant human memories have been carried to the present via folklore? I once attended the premier of a great film adaption of Beowulf by the Icelandic-Canadian director Sturla Gunnarsson (Beowulf & Grendel, 2005), stunningly shot in Iceland. I am still intrigued whether he was depicting Grendel as a solitary surviving Neanderthal. After the movie Gunnarsson fielded questions from the audience, but I was too chicken to pipe up, and never asked my question – damn!
From a scientific perspective H. floriensis presents many questions. Did they evolve from H. erectus, H. denisoviensis, H. neanderthalensis, or perhaps some combination? Is their size a very specific local adaption caused by the well-known process of island dwarfism, or did they fill a wide-ranging ecological niche and we simply haven’t yet found more remains elsewhere? Did they overlap with the first wave of H. sapiens to pass through the Indonesian archipelago? Asia has been generally underexplored and studied by paleoanthropologists, and I expect that it will produce many revelatory finds in the coming years.
Homo Sapiens
Just recently there has been a lot of debate about when and where H. sapiens first emerged as a distinct species. In June of 2017 a stunning discovery was published in Nature – H. sapiens remains found at the Jebel Irhoud site between Marrakesh, Morocco and the Atlantic coast, had been dated to 315,000 ya! Not only were the bones much older than previously expected for H. sapiens, they were in the wrong place, up in the NW corner of Africa rather than East Africa’s Rift Valley. This has driven a grand rethinking of prevailing theories. Rather than incubating in a relatively small East African pocket, scientists are now thinking that favourable climatic conditions around 300,000 ya (there’s that convenient glacial cycle again) drove a much more continental evolution of H. sapiens, as many different sub-pockets of the proto-species roamed around Africa and interbred. Also, the skull morphology suggests that in these early days of our species, it was the face that took on the characteristic modern look with a sharp jaw and much less pronounced browridge first, while the back of the skull retained its archaic elongated shape.
Just a month after that paper, another bombshell was dropped in the same journal, with details published on new work at a Northern Territory rock shelter site in Australia named Madjedbebe. It concluded that the first humans arrived there 65,000 ya, some 20,000 years earlier than previously thought. Not only does this mean the ancestors of today’s Australian Aboriginals overlapped with many of Australia’s ancient megafauna (and logically are implicated in their extinction), it also means that they passed right by the island of Flores while H. floriensis still existed. This earlier arrival in Australia has had implications on when humans first left Africa. This expansion likely occurred much earlier, to allow time for these ancient, primitive humans to gradually migrate from Africa all the way to Australia (allowing of course for inter-species sex stops along the way with Neanderthals and Denisovans). This is why I have used an estimated out-of-Africa date of ~100,000 ya rather than the commonly used 60,000 ya.
Paleoanthropology is a fascinating field, and advances in many areas of science are continually expanding our knowledge of our species’ prehistory and that of our Hominin relatives. Time relentlessly moves forward as well, and it is interesting to think that the forces of evolution will drive future splits and speciations. Perhaps during some dark age following the collapse of our current modern era, pockets of humans will be isolated and evolve into new Hominin species. And maybe at some point in the distant future history will repeat itself, and yet another world-conquering Hominin species will emerge from Africa, the 4th such great wave.










Share This Column